A04 - Data Wrangling#
Revised
19 Jun 2023
Programming Environment#
library(tidyverse) # includes lots of data verbs like `group_by()` and `summarise()`
library(mosaicData) # includes the `HELPmiss` data set
sessionInfo()
── Attaching core tidyverse packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.2 ✔ readr 2.1.4
✔ forcats 1.0.0 ✔ stringr 1.5.0
✔ ggplot2 3.4.3 ✔ tibble 3.2.1
✔ lubridate 1.9.2 ✔ tidyr 1.3.0
✔ purrr 1.0.2
── Conflicts ─────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
R version 4.3.0 (2023-04-21)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS 14.4.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/New_York
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] mosaicData_0.20.3 lubridate_1.9.2 forcats_1.0.0 stringr_1.5.0
[5] dplyr_1.1.2 purrr_1.0.2 readr_2.1.4 tidyr_1.3.0
[9] tibble_3.2.1 ggplot2_3.4.3 tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] gtable_0.3.3 jsonlite_1.8.5 compiler_4.3.0 crayon_1.5.2
[5] tidyselect_1.2.0 IRdisplay_1.1 scales_1.2.1 uuid_1.1-0
[9] fastmap_1.1.1 IRkernel_1.3.2 R6_2.5.1 generics_0.1.3
[13] munsell_0.5.0 pillar_1.9.0 tzdb_0.4.0 rlang_1.1.1
[17] utf8_1.2.3 stringi_1.7.12 repr_1.1.6 timechange_0.2.0
[21] cli_3.6.1 withr_2.5.0 magrittr_2.0.3 digest_0.6.31
[25] grid_4.3.0 base64enc_0.1-3 hms_1.1.3 pbdZMQ_0.3-9
[29] lifecycle_1.0.3 vctrs_0.6.3 evaluate_0.21 glue_1.6.2
[33] fansi_1.0.4 colorspace_2.1-0 tools_4.3.0 pkgconfig_2.0.3
[37] htmltools_0.5.5
# Load the `HELPmiss` data set into our RStudio environment
# data("HELPmiss", package = "mosaicData")
?mosaicData::HELPmiss
head(HELPmiss)
| age | anysub | cesd | d1 | daysanysub | dayslink | drugrisk | e2b | female | sex | ⋯ | pcs | pss_fr | racegrp | satreat | sexrisk | substance | treat | avg_drinks | max_drinks | hospitalizations | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <int> | <fct> | <int> | <int> | <int> | <int> | <int> | <int> | <int> | <fct> | ⋯ | <dbl> | <int> | <fct> | <fct> | <int> | <fct> | <fct> | <int> | <int> | <int> | |
| 1 | 37 | yes | 49 | 3 | 177 | 225 | 0 | NA | 0 | male | ⋯ | 58.41369 | 0 | black | no | 4 | cocaine | yes | 13 | 26 | 3 |
| 2 | 37 | yes | 30 | 22 | 2 | NA | 0 | NA | 0 | male | ⋯ | 36.03694 | 1 | white | no | 7 | alcohol | yes | 56 | 62 | 22 |
| 3 | 26 | yes | 39 | 0 | 3 | 365 | 20 | NA | 0 | male | ⋯ | 74.80633 | 13 | black | no | 2 | heroin | no | 0 | 0 | 0 |
| 4 | 39 | yes | 15 | 2 | 189 | 343 | 0 | 1 | 1 | female | ⋯ | 61.93168 | 11 | white | yes | 4 | heroin | no | 5 | 5 | 2 |
| 5 | 32 | yes | 39 | 12 | 2 | 57 | 0 | 1 | 0 | male | ⋯ | 37.34558 | 10 | black | no | 6 | cocaine | no | 10 | 13 | 12 |
| 6 | 47 | yes | 6 | 1 | 31 | 365 | 0 | NA | 1 | female | ⋯ | 46.47521 | 5 | black | no | 5 | cocaine | yes | 4 | 4 | 1 |
# number of cases
HELPmiss %>%
summarize(count = n())
| count |
|---|
| <int> |
| 470 |
# total e2b
HELPmiss %>%
summarize(
total_e2b = sum(e2b, na.rm = TRUE)
)
| total_e2b |
|---|
| <int> |
| 549 |
# mean daysanysub
HELPmiss %>%
summarize(
mean_daysanysub = mean(daysanysub, na.rm = TRUE)
)
| mean_daysanysub |
|---|
| <dbl> |
| 75.13095 |
HELPmiss %>%
group_by(sex) %>%
summarize(
count = n(),
total_e2b = sum(e2b, na.rm = TRUE),
mean_daysanysub = mean(daysanysub, na.rm = TRUE)
)
| sex | count | total_e2b | mean_daysanysub |
|---|---|---|---|
| <fct> | <int> | <int> | <dbl> |
| male | 359 | 457 | 72.60513 |
| female | 111 | 92 | 83.77193 |
HELPmiss %>%
group_by(homeless) %>%
summarize(
count = n(),
total_e2b = sum(e2b, na.rm = TRUE),
mean_daysanysub = mean(daysanysub, na.rm = TRUE)
)
| homeless | count | total_e2b | mean_daysanysub |
|---|---|---|---|
| <fct> | <int> | <int> | <dbl> |
| housed | 251 | 160 | 77.10078 |
| homeless | 219 | 389 | 73.06504 |
HELPmiss %>%
group_by(substance) %>%
summarize(
count = n(),
total_e2b = sum(e2b, na.rm = TRUE),
mean_daysanysub = mean(daysanysub, na.rm = TRUE)
)
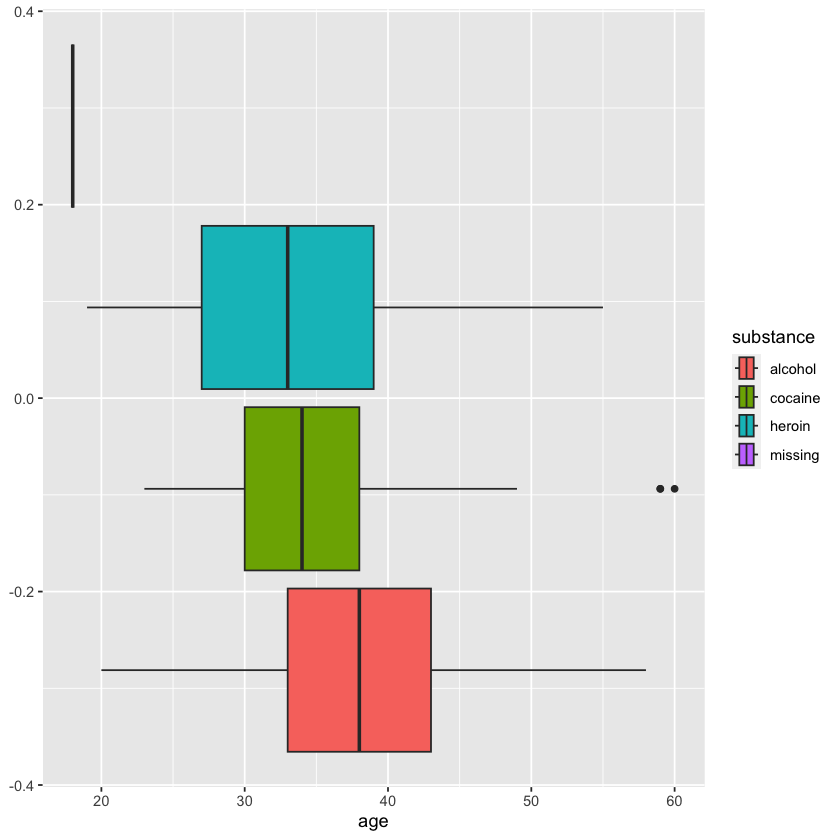
| substance | count | total_e2b | mean_daysanysub |
|---|---|---|---|
| <fct> | <int> | <int> | <dbl> |
| alcohol | 185 | 242 | 78.49495 |
| cocaine | 156 | 98 | 86.67033 |
| heroin | 128 | 208 | 52.82258 |
| missing | 1 | 1 | NaN |
HELPmiss %>%
group_by(homeless, sex) %>%
summarize(
count = n(),
total_e2b = sum(e2b, na.rm = TRUE),
mean_daysanysub = mean(daysanysub, na.rm = TRUE)
)
`summarise()` has grouped output by 'homeless'. You can override using the `.groups` argument.
| homeless | sex | count | total_e2b | mean_daysanysub |
|---|---|---|---|---|
| <fct> | <fct> | <int> | <int> | <dbl> |
| housed | male | 182 | 110 | 74.30851 |
| housed | female | 69 | 50 | 84.60000 |
| homeless | male | 177 | 347 | 71.01980 |
| homeless | female | 42 | 42 | 82.45455 |
HELPmiss %>%
group_by(homeless, substance) %>%
summarize(
count = n(),
total_e2b = sum(e2b, na.rm = TRUE),
mean_daysanysub = mean(daysanysub, na.rm = TRUE)
)
`summarise()` has grouped output by 'homeless'. You can override using the `.groups` argument.
| homeless | substance | count | total_e2b | mean_daysanysub |
|---|---|---|---|---|
| <fct> | <fct> | <int> | <int> | <dbl> |
| housed | alcohol | 76 | 34 | 104.79487 |
| housed | cocaine | 96 | 37 | 81.05556 |
| housed | heroin | 79 | 89 | 41.16667 |
| homeless | alcohol | 109 | 208 | 61.40000 |
| homeless | cocaine | 60 | 61 | 94.86486 |
| homeless | heroin | 49 | 119 | 68.96154 |
| homeless | missing | 1 | 1 | NaN |
HELPmiss %>%
count(substance) %>%
arrange(desc(n))
| substance | n |
|---|---|
| <fct> | <int> |
| alcohol | 185 |
| cocaine | 156 |
| heroin | 128 |
| missing | 1 |
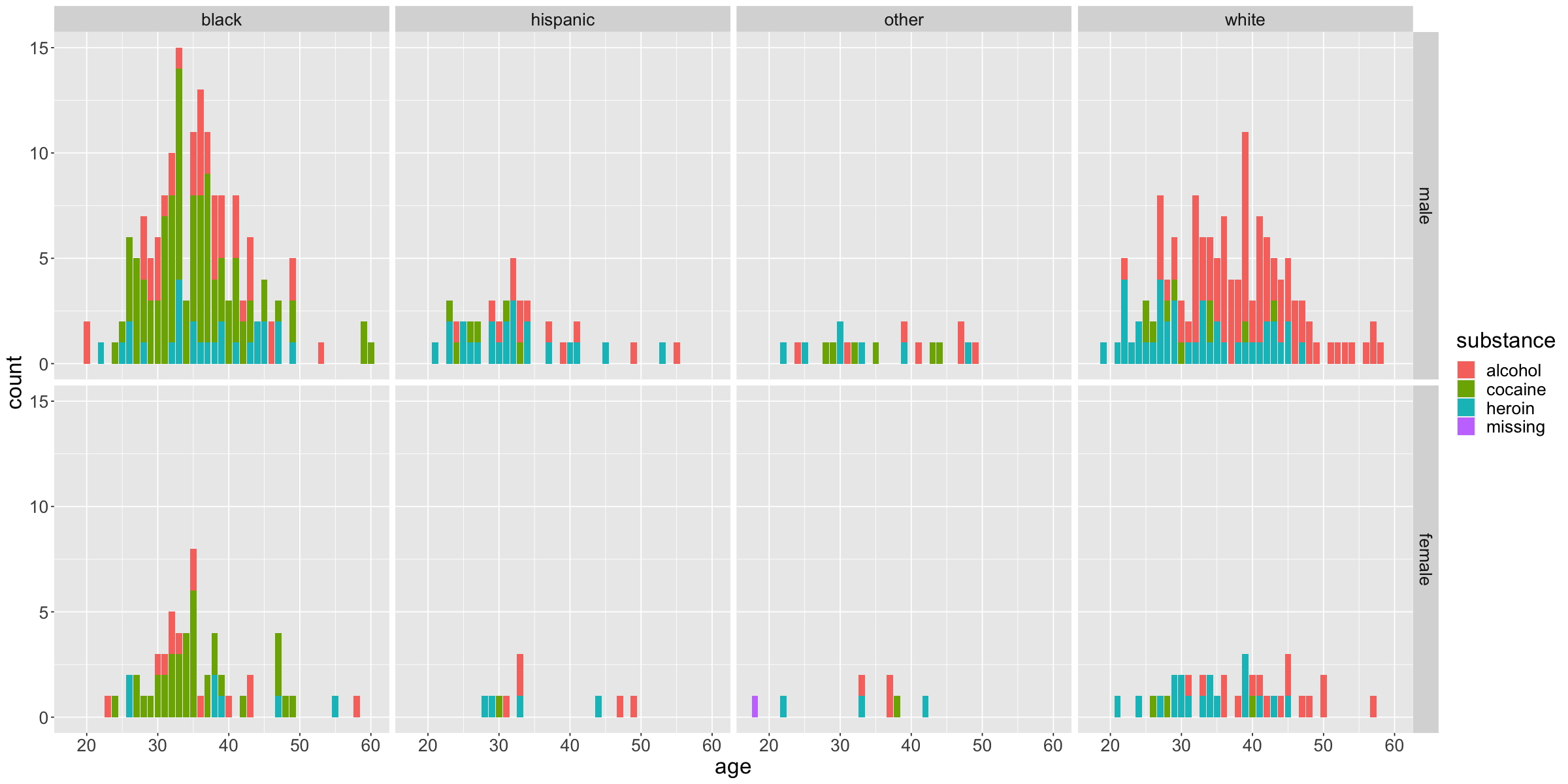
options(repr.plot.width = 20, repr.plot.height = 10)
ggplot(data = HELPmiss) +
geom_bar(
mapping = aes(x = age,
fill = substance),
position = 'stack',
show.legend = TRUE,
stat = 'count'
) +
facet_grid(sex ~ racegrp) +
theme(text = element_text(size = 20))