A10 - Base R#
Revised
20 Jun 2023
Programming Environment#
utils::sessionInfo()
R version 4.3.0 (2023-04-21)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS 15.2
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/New_York
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] digest_0.6.31 IRdisplay_1.1 utf8_1.2.3 base64enc_0.1-3
[5] fastmap_1.1.1 glue_1.6.2 htmltools_0.5.5 repr_1.1.6
[9] lifecycle_1.0.3 cli_3.6.1 fansi_1.0.4 vctrs_0.6.3
[13] pbdZMQ_0.3-9 compiler_4.3.0 tools_4.3.0 evaluate_0.21
[17] pillar_1.9.0 crayon_1.5.2 rlang_1.1.1 jsonlite_1.8.5
[21] IRkernel_1.3.2 uuid_1.1-0
Base R#
base::library(help = 'base')
base::library(help = 'utils')
base::library(help = 'datasets')
utils::head(iris)
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species | |
|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <fct> | |
| 1 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 2 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 3 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 4 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 5 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
| 6 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
Select the element in the 10th row and the 5th column of the iris data set.
datasets::iris[10, 5]
base::as.character(datasets::iris[10, 5])
base::as.factor(base::as.character(datasets::iris[10, 5]))
Levels:
- 'setosa'
- 'versicolor'
- 'virginica'
Levels:
'setosa'Select the species column of the iris data set and print the last five rows of that column.
class(iris$Species)
iris$Species
class(as.character(iris$Species))
as.character(iris$Species)
class(iris[['Species']])
iris[['Species']]
class(as.character(iris[['Species']]))
as.character(iris[['Species']])
class(iris['Species'])
iris['Species']
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
Levels:
- 'setosa'
- 'versicolor'
- 'virginica'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- setosa
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- versicolor
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
- virginica
Levels:
- 'setosa'
- 'versicolor'
- 'virginica'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'setosa'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'versicolor'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
| Species |
|---|
| <fct> |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| setosa |
| ⋮ |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
| virginica |
iris[(length(iris$Species) - 4):length(iris$Species),'Species']
as.character(iris[(length(iris$Species) - 4):length(iris$Species),'Species'])
tail(iris$Species, n = 5)
as.character(tail(iris$Species, n = 5))
tail(iris[['Species']], n = 5)
as.character(tail(iris[['Species']], n = 5))
tail(iris['Species'], n = 5)
- virginica
- virginica
- virginica
- virginica
- virginica
Levels:
- 'setosa'
- 'versicolor'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- virginica
- virginica
- virginica
- virginica
- virginica
Levels:
- 'setosa'
- 'versicolor'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- virginica
- virginica
- virginica
- virginica
- virginica
Levels:
- 'setosa'
- 'versicolor'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
- 'virginica'
| Species | |
|---|---|
| <fct> | |
| 146 | virginica |
| 147 | virginica |
| 148 | virginica |
| 149 | virginica |
| 150 | virginica |
What is the average sepal length of flowers with petal widths greater than one?
base::mean(iris$Sepal.Length[iris$Petal.Width > 1])
Write a for loop that iterates over the numbers 1 to 10. If the number is less than five, print the product of the number with 3; otherwise, print the product of the number with 2.
for (i in 1:10) {
if (i < 5) {
base::print(i * 3)
} else {
base::print(i * 2)
}
}
[1] 3
[1] 6
[1] 9
[1] 12
[1] 10
[1] 12
[1] 14
[1] 16
[1] 18
[1] 20
Write a while loop that prints out random normal numbers with mean equal to 1 and standard deviation equal to 2 (use rnorm()) but breaks for a number greater than 3.
base::set.seed(184)
num <- stats::rnorm(n = 1, mean = 1, sd = 2)
while (num < 3) {
base::print(num)
num <- stats::rnorm(n = 1, mean = 1, sd = 2)
}
[1] 0.1167419
[1] 1.588951
[1] 2.436941
[1] 0.932507
[1] 2.408507
[1] -1.206123
[1] -1.158457
base::set.seed(184)
while (TRUE) {
num <- stats::rnorm(n = 1, mean = 1, sd = 2)
if (num > 3) {
break
}
base::print(num)
}
[1] 0.1167419
[1] 1.588951
[1] 2.436941
[1] 0.932507
[1] 2.408507
[1] -1.206123
[1] -1.158457
Use a nested for loop to produce the following data frame. Name the data frame Counting and the variables V1, V2, V3, and V4.
V1 |
V2 |
V3 |
V4 |
|---|---|---|---|
1 |
2 |
3 |
4 |
2 |
3 |
4 |
5 |
3 |
4 |
5 |
6 |
Counting <- base::data.frame()
Rows <- 1:3
Cols <- 1:4
for (j in 1:base::length(Cols)) {
for (i in 1:base::length(Rows)) {
Counting[i, j] <- i + j - 1
}
}
Counting
| V1 | V2 | V3 | V4 | |
|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | 1 | 2 | 3 | 4 |
| 2 | 2 | 3 | 4 | 5 |
| 3 | 3 | 4 | 5 | 6 |
base::colnames(Counting) <- base::sub('V', 'X', base::colnames(Counting))
Counting
| X1 | X2 | X3 | X4 | |
|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | |
| 1 | 1 | 2 | 3 | 4 |
| 2 | 2 | 3 | 4 | 5 |
| 3 | 3 | 4 | 5 | 6 |
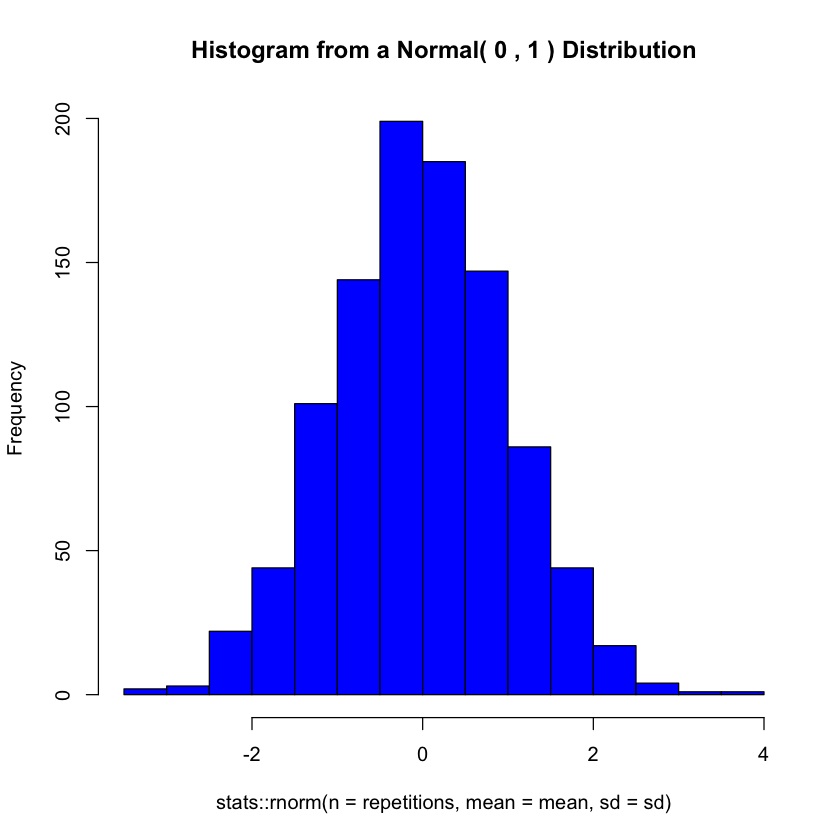
Complete the following user-defined function to generate n random normal numbers with a given mean and standard deviation and plot a histogram of it (use hist()). Call the function to generate 1000 random normal numbers with mean equal to 0 and standard deviation equal to 1 and plot its histogram.
base::set.seed(184)
plotNormal <- function (repetitions, mean, sd) {
graphics::hist(
col = 'blue',
main = base::paste('Histogram from a Normal(', mean, ',' , sd, ') Distribution'),
x = stats::rnorm(n = repetitions, mean = mean, sd = sd)
)
}
plotNormal(1000, 0, 1)
Vectors c(), :, seq(), rep()
Subsetting [] (vectors, data frames), [[]] (lists), $ (data frames, lists)
base::class(x = v)
utils::str(object = v)
base::summary(object = v)
# vector of doubles
v <- base::c(2, 5, NA, 5)
v
base::is.vector(v)
base::is.double(v)
base::is.atomic(v)
base::is.integer(v)
base::typeof(v)
base::mean(x = g)
base::mean(x = g, na.rm = TRUE)
- 2
- 5
- <NA>
- 5
Error in eval(expr, envir, enclos): object 'g' not found
Traceback:
1. base::mean(x = g)
# vector of integers
v <- base::c(2L, 5L, NA, 5L)
v
base::is.vector(v)
base::is.double(v)
base::is.atomic(v)
base::is.integer(v)
base::typeof(v)
base::mean(x = g)
base::mean(x = g, na.rm = TRUE)
- 2
- 5
- <NA>
- 5
# vector of integers
v <- 2:6
v
base::is.vector(v)
base::is.double(v)
base::is.atomic(v)
base::is.integer(v)
base::typeof(v)
- 2
- 3
- 4
- 5
- 6
# vector of doubles
v <- seq(2, 3, by = 0.2)
v
base::is.vector(v)
base::is.double(v)
base::is.atomic(v)
base::is.integer(v)
base::typeof(v)
- 2
- 2.2
- 2.4
- 2.6
- 2.8
- 3
# vector of doubles
v <- seq(2, 5, length.out = 5)
v
base::is.vector(v)
base::is.double(v)
base::is.atomic(v)
base::is.integer(v)
base::typeof(v)
- 2
- 2.75
- 3.5
- 4.25
- 5
# vector of characters
v <- rep(c('a', 'b', 'c'), times = 3)
v
base::is.vector(v)
base::is.double(v)
base::is.atomic(v)
base::is.integer(v)
base::is.character(v)
base::typeof(v)
- 'a'
- 'b'
- 'c'
- 'a'
- 'b'
- 'c'
- 'a'
- 'b'
- 'c'
# vector of characters
v <- rep(c('a', 'b', 'c'), each = 3)
v
base::is.vector(v)
base::is.double(v)
base::is.atomic(v)
base::is.integer(v)
base::is.character(v)
base::typeof(v)
- 'a'
- 'a'
- 'a'
- 'b'
- 'b'
- 'b'
- 'c'
- 'c'
- 'c'
v <- c('a', 'b', 'c', 1, 2, 3)
v
base::is.vector(v)
base::is.double(v)
base::is.atomic(v)
base::is.integer(v)
base::is.character(v)
base::typeof(v)
- 'a'
- 'b'
- 'c'
- '1'
- '2'
- '3'
v[4]
v[2:4]
v[c(1, 5)]
v[-4]
v[-c(1, 4, 6)]
- 'b'
- 'c'
- '1'
- 'a'
- '2'
- 'a'
- 'b'
- 'c'
- '2'
- '3'
- 'b'
- 'c'
- '2'
v1 <- c('a', 'b', 'c', 'd', 'e', 'f')
v2 <- 1:6
df <- base::data.frame(v1, v2)
df
| v1 | v2 |
|---|---|
| <chr> | <int> |
| a | 1 |
| b | 2 |
| c | 3 |
| d | 4 |
| e | 5 |
| f | 6 |
df[3, 2]
df[4,]
| v1 | v2 | |
|---|---|---|
| <chr> | <int> | |
| 4 | d | 4 |
df[,2]
- 1
- 2
- 3
- 4
- 5
- 6
base::set.seed(184)
y <- base::sample(v1, 10, replace = TRUE)
y
y == 'g'
y[y == 'g']
y[y %in% c('e', 'f', 'g')]
base::which(y %in% c('e', 'f', 'g'))
- 'c'
- 'f'
- 'f'
- 'f'
- 'f'
- 'a'
- 'f'
- 'e'
- 'f'
- 'c'
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- 'f'
- 'f'
- 'f'
- 'f'
- 'f'
- 'e'
- 'f'
- 2
- 3
- 4
- 5
- 7
- 8
- 9
numbers <- 1:26
letters <- base::letters
df <- base::data.frame(numbers, letters)
df
| numbers | letters |
|---|---|
| <int> | <chr> |
| 1 | a |
| 2 | b |
| 3 | c |
| 4 | d |
| 5 | e |
| 6 | f |
| 7 | g |
| 8 | h |
| 9 | i |
| 10 | j |
| 11 | k |
| 12 | l |
| 13 | m |
| 14 | n |
| 15 | o |
| 16 | p |
| 17 | q |
| 18 | r |
| 19 | s |
| 20 | t |
| 21 | u |
| 22 | v |
| 23 | w |
| 24 | x |
| 25 | y |
| 26 | z |
df$numbers
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
df$combo <- base::paste(letters, '-', numbers, 'combo')
df$uppercase <- base::LETTERS
df$states <- NA
for (i in 1:base::nrow(df)) {
df$states[i] <- datasets::state.abb[i]
}
df
| numbers | letters | combo | uppercase | states |
|---|---|---|---|---|
| <int> | <chr> | <chr> | <chr> | <chr> |
| 1 | a | a - 1 combo | A | AL |
| 2 | b | b - 2 combo | B | AK |
| 3 | c | c - 3 combo | C | AZ |
| 4 | d | d - 4 combo | D | AR |
| 5 | e | e - 5 combo | E | CA |
| 6 | f | f - 6 combo | F | CO |
| 7 | g | g - 7 combo | G | CT |
| 8 | h | h - 8 combo | H | DE |
| 9 | i | i - 9 combo | I | FL |
| 10 | j | j - 10 combo | J | GA |
| 11 | k | k - 11 combo | K | HI |
| 12 | l | l - 12 combo | L | ID |
| 13 | m | m - 13 combo | M | IL |
| 14 | n | n - 14 combo | N | IN |
| 15 | o | o - 15 combo | O | IA |
| 16 | p | p - 16 combo | P | KS |
| 17 | q | q - 17 combo | Q | KY |
| 18 | r | r - 18 combo | R | LA |
| 19 | s | s - 19 combo | S | ME |
| 20 | t | t - 20 combo | T | MD |
| 21 | u | u - 21 combo | U | MA |
| 22 | v | v - 22 combo | V | MI |
| 23 | w | w - 23 combo | W | MN |
| 24 | x | x - 24 combo | X | MS |
| 25 | y | y - 25 combo | Y | MO |
| 26 | z | z - 26 combo | Z | MT |
for (i in 1:4) {
j <- i + 10
base::print(j)
}
[1] 11
[1] 12
[1] 13
[1] 14
byFives <- base::seq(0, 100, by = 5)
byFives
for (i in 1:base::length(byFives)) {
j <- byFives[i] / 3
base::print(j)
}
- 0
- 5
- 10
- 15
- 20
- 25
- 30
- 35
- 40
- 45
- 50
- 55
- 60
- 65
- 70
- 75
- 80
- 85
- 90
- 95
- 100
[1] 0
[1] 1.666667
[1] 3.333333
[1] 5
[1] 6.666667
[1] 8.333333
[1] 10
[1] 11.66667
[1] 13.33333
[1] 15
[1] 16.66667
[1] 18.33333
[1] 20
[1] 21.66667
[1] 23.33333
[1] 25
[1] 26.66667
[1] 28.33333
[1] 30
[1] 31.66667
[1] 33.33333
numbers <- base::seq(10, 37, by = 1)
numbers
DivisibleBy5 <- rep(NA, length(numbers))
DivisibleBy5
for (i in 1:base::length(numbers)) {
remainder <- numbers[i] %% 5
if (remainder == 0) {
DivisibleBy5[i] <- TRUE
} else {
DivisibleBy5[i] <- FALSE
}
}
# ALTERNATIVE APPROACH
# index5 = 1
# for (i in numbers) {
# remainder <- i %% 5
# if (remainder == 0) {
# DivisibleBy5[index5] <- TRUE
# } else {
# DivisibleBy5[index5] <- FALSE
# }
# index5 <- index5 + 1
# }
DivisibleBy5
numbers[DivisibleBy5]
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- <NA>
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- 10
- 15
- 20
- 25
- 30
- 35
numbers %% 5 == 0
numbers[numbers %% 5 == 0]
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- TRUE
- FALSE
- FALSE
- 10
- 15
- 20
- 25
- 30
- 35
for.loop <- DivisibleBy5
vectorization <- numbers %% 5 == 0
base::sum(for.loop != vectorization) # = 0
df <- data.frame()
Rows <- 1:6
Cols <- 1:5
for (j in 1:base::length(Cols)) {
for (i in 1:base::length(Rows)) {
df[i, j] <- paste('Row', i, ' Col', j)
}
}
df
| V1 | V2 | V3 | V4 | V5 | |
|---|---|---|---|---|---|
| <chr> | <chr> | <chr> | <chr> | <chr> | |
| 1 | Row 1 Col 1 | Row 1 Col 2 | Row 1 Col 3 | Row 1 Col 4 | Row 1 Col 5 |
| 2 | Row 2 Col 1 | Row 2 Col 2 | Row 2 Col 3 | Row 2 Col 4 | Row 2 Col 5 |
| 3 | Row 3 Col 1 | Row 3 Col 2 | Row 3 Col 3 | Row 3 Col 4 | Row 3 Col 5 |
| 4 | Row 4 Col 1 | Row 4 Col 2 | Row 4 Col 3 | Row 4 Col 4 | Row 4 Col 5 |
| 5 | Row 5 Col 1 | Row 5 Col 2 | Row 5 Col 3 | Row 5 Col 4 | Row 5 Col 5 |
| 6 | Row 6 Col 1 | Row 6 Col 2 | Row 6 Col 3 | Row 6 Col 4 | Row 6 Col 5 |