A06 - Project: Bird Species#
Kaplan, Daniel & Matthew Beckman. (2021). Data Computing. 2nd Ed. Home.
Revised
19 Jun 2023
Programming Environment#
library(dcData)
library(tidyverse)
sessionInfo()
── Attaching core tidyverse packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.2 ✔ readr 2.1.4
✔ forcats 1.0.0 ✔ stringr 1.5.0
✔ ggplot2 3.4.3 ✔ tibble 3.2.1
✔ lubridate 1.9.2 ✔ tidyr 1.3.0
✔ purrr 1.0.2
── Conflicts ─────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
R version 4.3.0 (2023-04-21)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS 14.4.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/New_York
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] lubridate_1.9.2 forcats_1.0.0 stringr_1.5.0 dplyr_1.1.2
[5] purrr_1.0.2 readr_2.1.4 tidyr_1.3.0 tibble_3.2.1
[9] ggplot2_3.4.3 tidyverse_2.0.0 dcData_0.1.0
loaded via a namespace (and not attached):
[1] gtable_0.3.3 jsonlite_1.8.5 compiler_4.3.0 crayon_1.5.2
[5] tidyselect_1.2.0 IRdisplay_1.1 scales_1.2.1 uuid_1.1-0
[9] fastmap_1.1.1 IRkernel_1.3.2 R6_2.5.1 generics_0.1.3
[13] munsell_0.5.0 pillar_1.9.0 tzdb_0.4.0 rlang_1.1.1
[17] utf8_1.2.3 stringi_1.7.12 repr_1.1.6 timechange_0.2.0
[21] cli_3.6.1 withr_2.5.0 magrittr_2.0.3 digest_0.6.31
[25] grid_4.3.0 base64enc_0.1-3 hms_1.1.3 pbdZMQ_0.3-9
[29] lifecycle_1.0.3 vctrs_0.6.3 evaluate_0.21 glue_1.6.2
[33] fansi_1.0.4 colorspace_2.1-0 tools_4.3.0 pkgconfig_2.0.3
[37] htmltools_0.5.5
Get the data#
?dcData::OrdwayBirds
?dcData::OrdwaySpeciesNames
OrdwayBirds %>%
select(Month, Day) %>%
head()
| Month | Day | |
|---|---|---|
| <chr> | <chr> | |
| 3 | 7 | 16 |
| 4 | ||
| 5 | 7 | 16 |
| 6 | 7 | 16 |
| 7 | 7 | 16 |
| 8 | 7 | 16 |
OrdwayBirds <-
OrdwayBirds %>%
select(SpeciesName, Month, Day) %>%
mutate(
Month = as.numeric(as.character(Month)),
Day = as.numeric(as.character(Day))
)
OrdwayBirds %>%
head()
| SpeciesName | Month | Day | |
|---|---|---|---|
| <chr> | <dbl> | <dbl> | |
| 3 | Song Sparrow | 7 | 16 |
| 4 | NA | NA | |
| 5 | Song Sparrow | 7 | 16 |
| 6 | Field Sparrow | 7 | 16 |
| 7 | Field Sparrow | 7 | 16 |
| 8 | Song Sparrow | 7 | 16 |
OrdwaySpeciesNames %>%
filter(is.na(SpeciesNameCleaned))
| SpeciesName | SpeciesNameCleaned |
|---|---|
| <chr> | <chr> |
| NA | |
| -lost- | NA |
| -missing- | NA |
| [Nothing, just dashes] | NA |
| 13:00:00 | NA |
| Bank Swallow | NA |
| Barn Swallow | NA |
| Bay-breasted Warbler | NA |
| Blackpoll Warbler | NA |
| Blue Jay | NA |
| Blue-headed Vireo | NA |
| Blue-winged Warbler | NA |
| Bluebird | NA |
| Boreal Chickadee | NA |
| Brewer's Sparrow | NA |
| Brown Creeper | NA |
| Brown Thrasher | NA |
| Brown Towhee | NA |
| Cactus Wren | NA |
| Common Crow | NA |
| Common Grackle | NA |
| Common Nighthawk | NA |
| Common Redpoll | NA |
| Common Yellowthroat | NA |
| Connecticut Warbler | NA |
| Downy Woodpecker | NA |
| E Bluebird | NA |
| Eastern Bluebird | NA |
| Eastern Kingbird | NA |
| Eastern Meadowlark | NA |
| Eastern Robin | NA |
| Flicker | NA |
| Fox Sparrow | NA |
| Goldfinch | NA |
| Grackle | NA |
| Green Heron | NA |
| Ground Dove | NA |
| Hairy Woodpecker | NA |
| Hermit Thrush | NA |
| Horned Lark | NA |
| House Finch | NA |
| House Sparrow | NA |
| Inca Dove | NA |
| Indigo Bunting | NA |
| Killdeer | NA |
| Kingbird | NA |
| Kiskadee F.C. | NA |
| Magnolia Warbler | NA |
| Mockingbird | NA |
| Rough-winged Swallow | NA |
Task 1#
[1] Including misspellings, how many different species are there in the OrdwayBirds data?
There are 275 unique values of the variable SpeciesName. This reduces to 268 after dropping the following invalid values:
'''-lost-''-missing-''13:00:00''[Nothing, just dashes]''lost''none'
[2] Consider the OrdwaySpeciesNames data frame also found in the dcData package as well. How many distinct species are there in the SpeciesNameCleaned variable in OrdwaySpeciesNames? You will find it helpful to use n_distinct() a reduction function, which counts the number of unique values in a variable.
There are 108 unique values of the variable SpeciesNameCleaned after accounting for the value NA.
OrdwayBirds %>%
count(SpeciesName)
| SpeciesName | n |
|---|---|
| <chr> | <int> |
| 4 | |
| -lost- | 1 |
| -missing- | 1 |
| 13:00:00 | 1 |
| Acadian Flycatcher | 1 |
| American Gold Finch | 50 |
| American Goldfinch | 1153 |
| American Golf Finch | 1 |
| American Redfinch | 1 |
| American Redstart | 3 |
| American Robin | 4 |
| Arkansas Kingbird | 1 |
| Baltimore Oriole | 206 |
| Bank Swallow | 21 |
| Barn Swallow | 23 |
| Batimore Oriole | 1 |
| Bay-breasted Warbler | 2 |
| Blac-capped Chickadee | 1 |
| Black and White Warbler | 9 |
| Black-Capped Chickadee | 13 |
| Black-and-white Warbler | 1 |
| Black-billed Cookoo | 1 |
| Black-billed Cuckoo | 15 |
| Black-capeed Chickadee | 1 |
| Black-capped Chicakdee | 1 |
| Black-capped Chickadee | 1110 |
| Black-capped Chikadee | 1 |
| Black-capped chickadee | 187 |
| Black-throat Sparrow | 31 |
| Black-throat-Sparrow | 1 |
| ⋮ | ⋮ |
| White-breast Nuthatch | 23 |
| White-breasted Nuthatch | 236 |
| White-crown Sparrow | 17 |
| White-crowned Sparrow | 78 |
| White-eyed Vireo | 1 |
| White-thorat Sparrow | 1 |
| White-throat Sparrow | 86 |
| White-throated Sparrow | 229 |
| White-winged Junco | 2 |
| Wht-brstd Nuthatch | 1 |
| Wilson Warbler | 4 |
| Wilson's Warbler | 22 |
| Winter Wren | 1 |
| Wood Pewee | 37 |
| Wood Thrush | 3 |
| Woodcock | 1 |
| Wren | 2 |
| Yellow Flicker | 1 |
| Yellow Shafted Flicker | 4 |
| Yellow Warbler | 19 |
| Yellow-bellied Flycatcher | 7 |
| Yellow-bellied Sapsucker | 3 |
| Yellow-shaft Flicker | 6 |
| Yellow-shafted Flicker | 34 |
| Yellow-shafted flicker | 6 |
| Yellow-tailed Oriole | 1 |
| Yellowthroat | 107 |
| [Nothing, just dashes] | 1 |
| lost | 1 |
| none | 2 |
OrdwayBirds %>%
select(SpeciesName) %>%
n_distinct()
OrdwaySpeciesNames %>%
count(SpeciesNameCleaned)
| SpeciesNameCleaned | n |
|---|---|
| <chr> | <int> |
| Acadian Flycatcher | 1 |
| American Goldfinch | 3 |
| American Redfinch | 1 |
| American Redstart | 1 |
| American Robin | 1 |
| Arkansas Kingbird | 1 |
| Baltimore Oriole | 3 |
| Black and White Warbler | 2 |
| Black-billed Cookoo | 2 |
| Black-capped Chickadee | 8 |
| Black-throat Sparrow | 2 |
| Brown-headed Cowbird | 2 |
| Cardinal | 2 |
| Carolina Chickadee | 1 |
| Catbird | 4 |
| Cedar Waxwing | 2 |
| Chestnut-backed Chickadee | 1 |
| Chestnut-sided Warbler | 1 |
| Chickadee | 2 |
| Chipping Sparrow | 4 |
| Clay-colored Sparrow | 3 |
| Cowbird | 1 |
| Curve-billed Thrasher | 2 |
| Eastern Phoebe | 2 |
| Eastern Wood Pewee | 2 |
| Field Sparrow | 2 |
| Golden-Crowned Kinglet | 3 |
| Gray - cheeked Thrush | 4 |
| Great Crested Flycatcher | 3 |
| Harris's Sparrow | 3 |
| ⋮ | ⋮ |
| Tennessee Warbler | 2 |
| Traill's Flycatcher | 1 |
| Tree L | 1 |
| Tree Swallow | 4 |
| Tufted Titmouse | 1 |
| Unknown | 1 |
| Varied Thrush | 1 |
| Veery | 1 |
| Vesper Sparrow | 1 |
| Warbling Vireo | 1 |
| White-Crested Sparrow | 1 |
| White-Fronted Dove | 1 |
| White-breasted Nuthatch | 5 |
| White-crowned Sparrow | 2 |
| White-eyed Vireo | 1 |
| White-throat Sparrow | 5 |
| White-winged Junco | 1 |
| Wilson's Warbler | 2 |
| Winter Wren | 1 |
| Wood Pewee | 1 |
| Wood Thrush | 1 |
| Woodcock | 1 |
| Wren | 1 |
| Yellow Shafted Flicker | 5 |
| Yellow Warbler | 1 |
| Yellow-bellied Flycatcher | 1 |
| Yellow-bellied Sapsucker | 1 |
| Yellow-tailed Oriole | 1 |
| Yellowthroat | 1 |
| NA | 50 |
OrdwaySpeciesNames %>%
select(SpeciesNameCleaned) %>%
n_distinct()
Task 2#
Use the OrdwaySpeciesNames table to create a new data frame that corrects the misspellings in SpeciesNames. This can be done easily using the inner_join() data verb. Look at the names of the variables in OrdwaySpeciesNames and OrdwayBirds.
[1] Which variable(s) was used for matching cases?
The variable SpeciesName was used for matching cases.
[2] What were the variable(s) that will be added?
The variables SpeciesNameCleaned (renamed to Species), Month, and Day will be added.
Corrected <-
OrdwayBirds %>%
inner_join(y = OrdwaySpeciesNames) %>%
select(Species = SpeciesNameCleaned, Month, Day) %>%
na.omit()
Corrected %>%
head()
Joining with `by = join_by(SpeciesName)`
Warning message in inner_join(., y = OrdwaySpeciesNames):
“Detected an unexpected many-to-many relationship between `x` and `y`.
ℹ Row 4 of `x` matches multiple rows in `y`.
ℹ Row 211 of `y` matches multiple rows in `x`.
ℹ If a many-to-many relationship is expected, set `relationship = "many-to-many"` to silence this warning.”
| Species | Month | Day | |
|---|---|---|---|
| <chr> | <dbl> | <dbl> | |
| 1 | Song Sparrow | 7 | 16 |
| 3 | Song Sparrow | 7 | 16 |
| 4 | Field Sparrow | 7 | 16 |
| 5 | Field Sparrow | 7 | 16 |
| 6 | Field Sparrow | 7 | 16 |
| 7 | Field Sparrow | 7 | 16 |
Task 3#
Call the variable that contains the total count. Arrange this into descending order from the species with the most birds, and look through the list. (Hint: Remember n(). Also, one of the arguments to one of the data verbs will be desc(count) to arrange the cases into descending order. Display the top 10 species in terms of the number of bird captures.) Define for yourself a “major species” as a species with more than a particular threshold count. Set your threshold so that there are 5 or 6 species designated as major. Filter to produce a data frame with only the birds that belong to a major species. Save the output in a table called Majors. (Hint: Remember that summary functions can be used case-by-case when filtering or mutating a data frame that has been grouped.)
[1] How many bird captures are reported for each of the corrected species?
See below for the result (major species threshold >= 1000).
Corrected %>%
count(Species) %>%
arrange(desc(n)) %>%
head(n = 10)
| Species | n | |
|---|---|---|
| <chr> | <int> | |
| 1 | Slate-colored Junco | 2732 |
| 2 | Tree Swallow | 1537 |
| 3 | Black-capped Chickadee | 1327 |
| 4 | American Goldfinch | 1204 |
| 5 | Field Sparrow | 1164 |
| 6 | Lincoln's Sparrow | 790 |
| 7 | Robin | 608 |
| 8 | Catbird | 554 |
| 9 | Song Sparrow | 512 |
| 10 | House Wren | 460 |
Corrected %>%
group_by(Species) %>%
summarize(count = n()) %>%
arrange(desc(count)) %>%
head(n = 10)
| Species | count |
|---|---|
| <chr> | <int> |
| Slate-colored Junco | 2732 |
| Tree Swallow | 1537 |
| Black-capped Chickadee | 1327 |
| American Goldfinch | 1204 |
| Field Sparrow | 1164 |
| Lincoln's Sparrow | 790 |
| Robin | 608 |
| Catbird | 554 |
| Song Sparrow | 512 |
| House Wren | 460 |
Majors <-
Corrected %>%
group_by(Species) %>%
summarize(count = n()) %>%
arrange(desc(count)) %>%
filter(count >= 1000)
Majors
| Species | count |
|---|---|
| <chr> | <int> |
| Slate-colored Junco | 2732 |
| Tree Swallow | 1537 |
| Black-capped Chickadee | 1327 |
| American Goldfinch | 1204 |
| Field Sparrow | 1164 |
Task 4#
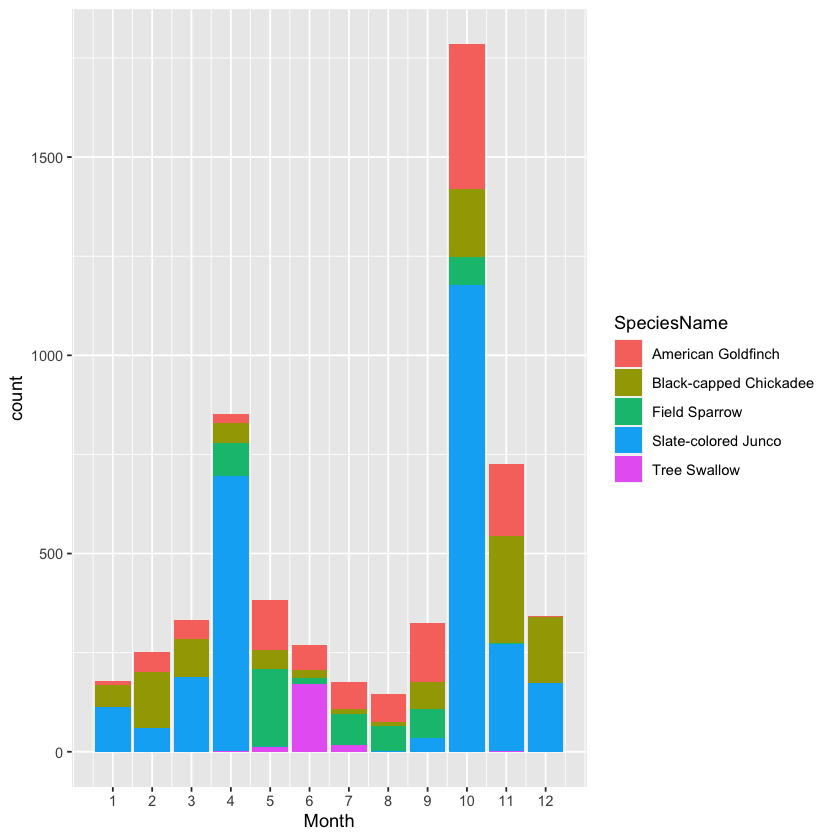
When you have correctly produced Majors, write a command that produces the month-by-month count of each of the major species. Call this table ByMonth. Display this month-by-month count with a bar chart arranged in a way that you think tells the story of what time of year the various species appear. You can use mplot() to explore different possibilies. (Warning: mplot() and similar interactive functions should not appear in your Rmd file, it needs to be used interactively from the console. Use the “Show Expression” button in mplot() to create an expression that you can cut and paste into a chunk in your Rmd document, so that the graph gets created when you compile it.) Once you have the graph, use it to answer these questions:
[1] Which species are present year-round?
American Goldfinch (11-12 mo)
Black-capped Chickadee (12 mo)
[2] Which species are migratory, that is, primarily present in one or two seasons?
Field Sparrow (6 mo)
Slate-colored Junco (8-9 mo)
Tree Swallow (3-5 mo)
[3] What is the peak month for each major species?
10American Goldfinch11Black-capped Chickadee05Field Sparrow10Slate-colored Junco06Tree Swallow
[4] Which major species are seen in good numbers for at least 6 months of the year? (Hint: n_distinct() and >= 6.)
Arguably, the only species that is not seen in good numbers for at least 6 months of the year is the tree swallow.
ByMonth <-
OrdwayBirds %>%
group_by(SpeciesName, Month = as.integer(Month)) %>%
summarize(count = n()) %>%
filter(SpeciesName %in% Majors$Species)
ByMonth
`summarise()` has grouped output by 'SpeciesName'. You can override using the `.groups` argument.
| SpeciesName | Month | count |
|---|---|---|
| <chr> | <int> | <int> |
| American Goldfinch | 1 | 10 |
| American Goldfinch | 2 | 51 |
| American Goldfinch | 3 | 48 |
| American Goldfinch | 4 | 21 |
| American Goldfinch | 5 | 125 |
| American Goldfinch | 6 | 63 |
| American Goldfinch | 7 | 67 |
| American Goldfinch | 8 | 70 |
| American Goldfinch | 9 | 151 |
| American Goldfinch | 10 | 364 |
| American Goldfinch | 11 | 180 |
| American Goldfinch | 12 | 3 |
| Black-capped Chickadee | 1 | 56 |
| Black-capped Chickadee | 2 | 140 |
| Black-capped Chickadee | 3 | 96 |
| Black-capped Chickadee | 4 | 51 |
| Black-capped Chickadee | 5 | 48 |
| Black-capped Chickadee | 6 | 20 |
| Black-capped Chickadee | 7 | 13 |
| Black-capped Chickadee | 8 | 11 |
| Black-capped Chickadee | 9 | 66 |
| Black-capped Chickadee | 10 | 173 |
| Black-capped Chickadee | 11 | 271 |
| Black-capped Chickadee | 12 | 165 |
| Field Sparrow | 4 | 83 |
| Field Sparrow | 5 | 197 |
| Field Sparrow | 6 | 15 |
| Field Sparrow | 7 | 79 |
| Field Sparrow | 8 | 64 |
| Field Sparrow | 9 | 74 |
| Field Sparrow | 10 | 69 |
| Field Sparrow | 11 | 1 |
| Slate-colored Junco | 1 | 113 |
| Slate-colored Junco | 2 | 61 |
| Slate-colored Junco | 3 | 188 |
| Slate-colored Junco | 4 | 694 |
| Slate-colored Junco | 5 | 1 |
| Slate-colored Junco | 8 | 1 |
| Slate-colored Junco | 9 | 35 |
| Slate-colored Junco | 10 | 1178 |
| Slate-colored Junco | 11 | 272 |
| Slate-colored Junco | 12 | 174 |
| Tree Swallow | 4 | 2 |
| Tree Swallow | 5 | 11 |
| Tree Swallow | 6 | 171 |
| Tree Swallow | 7 | 16 |
| Tree Swallow | 11 | 1 |
ByMonth %>%
group_by(SpeciesName) %>%
summarize(
MonthsPerYear = n(),
SixMonthsOrMore = n_distinct(Month) >= 6
)
| SpeciesName | MonthsPerYear | SixMonthsOrMore |
|---|---|---|
| <chr> | <int> | <lgl> |
| American Goldfinch | 12 | TRUE |
| Black-capped Chickadee | 12 | TRUE |
| Field Sparrow | 8 | TRUE |
| Slate-colored Junco | 10 | TRUE |
| Tree Swallow | 5 | FALSE |
ByMonth %>%
ggplot() +
geom_bar(
mapping = aes(x = Month, y = count, fill = SpeciesName),
na.rm = FALSE,
position = 'stack',
show.legend = TRUE,
stat = 'identity'
) +
scale_x_continuous(breaks = 1:12)